Todo ser humano está habitado por una serie de microorganismos que conforman lo que se conoce como microbioma humano. Tales microorganismos son necesarios para conservar la salud.
Al oír hablar de bacterias, virus y hongos, a todos nos suena a enfermedades. Sin embargo, se encuentran en los intestinos, los pulmones, genitales y la vejiga. Incluso, están en zonas como la boca, la nariz, la faringe y en el área cutánea.
Es tan importante la presencia de estos microorganismos en el cuerpo humano que hoy se sabe que aproximadamente la mitad de sus células son microbios. Mitad humano y mitad conformación bacteriana, el organismo es el resultado de la interdependencia entre ambos tipos de células, las cuales se relacionan activamente.
No obstante, no faltan estudios que afirman que el número de estos microorganismos supera grandemente al número de células humanas. Esta comunidad microbiana y bacteriana es lo que se conoce como microbioma.
Tabla de contenidos
Factores que intervienen en la formación del microbioma humano
Dependiendo de la zona del cuerpo, los microorganismos son diferentes. Así, la mayor carga se encuentra en los intestinos y en la boca, en menor grado a nivel cutáneo y vaginal.
Estas especies bacterianas que habitan en el cuerpo humano se encuentran por miles, son una gran variedad cuya amenaza para la salud del mismo es mínima. La composición de estos microorganismos varía en cada individuo, así como su evolución a lo largo del ciclo vital.
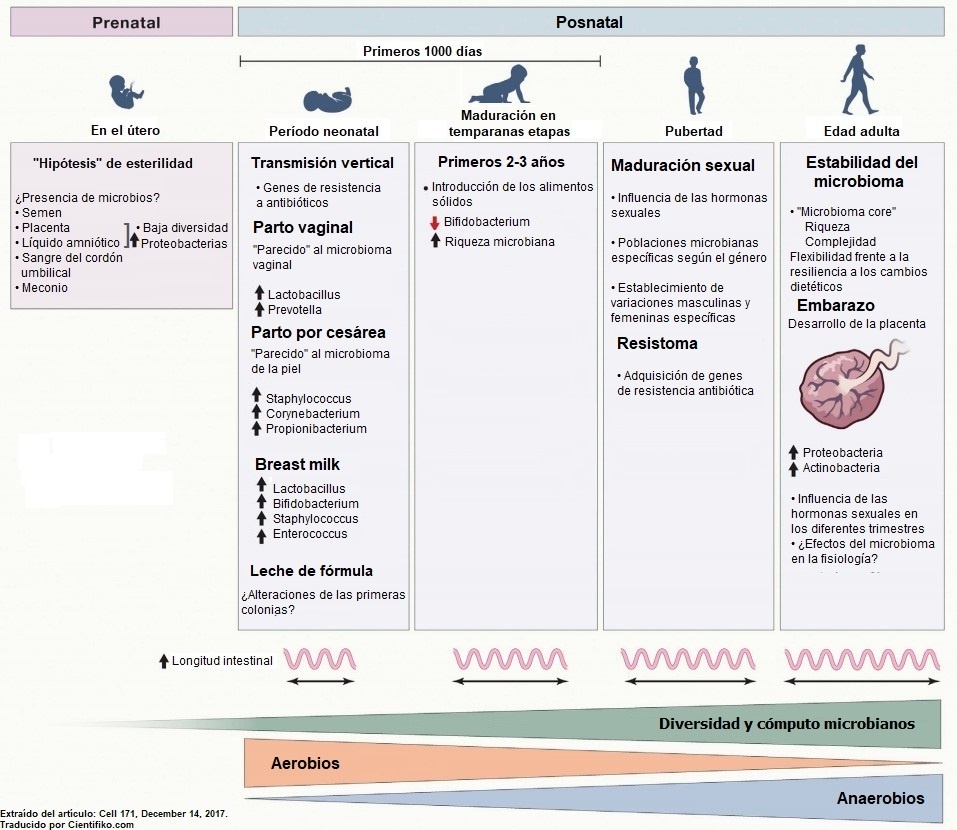
El ecosistema microbiano del cuerpo humanose va formando desde el momento del nacimiento, ya que depende de cómo se produjo, de la alimentación recibida y del ambiente. El tener contacto con animales domésticos o haber consumido antibióticos son también factores influyentes en el ecosistema de cada individuo.
El nacimiento y el Microbioma humano
Desde el nacimiento se tiene contacto con los microbios que proceden de la madre. Anteriormente se creía que los niños los adquieren en el momento del parto al pasar por el canal vaginal y luego al tener contacto con el ambiente.
Sin embargo, hoy se sabe que hay bacterias en la placenta, en el líquido amniótico y hasta en el cordón umbilical. Es así como se concluye que muchos microbios son herencia de la madre.
La manera en que se viene al mundo también influye en el ecosistema microbiano de cada individuo. Estudios han comprobado que los microbios intestinales de un niño que ha nacido por cesárea son parecidos a los de la piel de la madre. En cambio, en los nacidos por parto natural se encuentran bacterias propias de la vagina.
Asimismo, será diferente el sistema de un niño prematuro de aquel que nace de un embarazo llegado a término. Los niños alimentados con leche materna poseen bacterias y bacilos que no se encuentran en los niños que no se alimentan del pecho de sus madres. Es significativo que en las heces del niño se encuentren bacterias similares a las de la leche o a las de la piel de la progenitora.
Evolución del Microbioma humano
Es normal que el ecosistema microbiano evolucione con el paso del tiempo. Los recién nacidos tienen un microbioma bajo y cambiante de acuerdo a la alimentación y al ambiente, como ya se dijo antes. A medida que el niño crece el ecosistema se diversifica, aumentando en número y en variedad de un individuo a otro, debido a la maduración del organismo.
Durante los primeros años el sistema sigue acusando cambios, ya que intervienen las enfermedades infantiles, los antibióticos y los cambios en la dieta. Luego, con el arribo de la adolescencia, las hormonas también son factor de cambio, produciendo alteraciones que se manifiestan en la vida adulta. Todo ello se verá reflejado en la salud durante la mayoría de edad.
En los adultos los microbios se multiplican, diversifican y se hacen más estables y difíciles de cambiar. En cambio, durante la vejez los microbios disminuyen en número y variedad, a la vez que asemejan a los individuos entre sí.
Hay que resaltar que la genética interviene en todas estas variaciones, así como el sexo, el lugar de nacimiento y residencia, la ocupación y las relaciones con los congéneres.
La influencia del ecosistema microbiano en el organismo
La influencia más importante es la de los aportes nutricionales y la defensa contra las enfermedades. Los microbios que habitan los intestinos, por ejemplo, se encargan de metabolizar la bilis, las proteínas, así como las sustancias tóxicas o los medicamentos. A la vez producen vitaminas y ácidos que son fuentes de energía para las células humanas.
Otra función importante del ecosistema microbiano es que evita la presencia de otros microorganismos perjudiciales bloqueándolos en los intestinos. El sistema inmune también se ve afectado por la carga microbiana, así como las funciones del cerebro y del genoma humano. De tal manera que al influir en estos tres factores los microbios pueden determinar la conducta de los individuos.
También, se ha demostrado que los microbios pueden jugar un papel destacado en las funciones antinflamatorias de las células. A la vez que evitan las alergias y enfermedades como la diabetes, el lupus o la artritis reumatoide.
El ecosistema actúa defendiendo al organismo contra los agentes desencadenantes de enfermedades e impulsando la acción de los microbios propios. Para ello se establece una transacción entre las células humanas y las microbianas.
La influencia microbiana en las funciones cerebrales
La personalidad y las reacciones emocionales de todo ser humano están determinadas por la función cerebral. La carga microbiana también participa de ciertas pulsiones nerviosas que tienen que ver con la conducta de las personas y en los trastornos mentales.
Algunos estudios han comprobado estos efectos en ciertos procesos de construcción de conocimiento, en el desenvolvimiento social y, sobre todo, en los estados de ánimo.
El desarrollo de las neuronas y de enfermedades degenerativas también se ve condicionado por el sistema en cuestión. Los microbios del intestino se relacionan con las funciones del sistema nervioso. Por esa influencia hay quien ha llamado segundo cerebro a la función intestinal. Todo esto se conoce porque se ha comprobado que ciertas bacterias intestinales pueden producir neurotransmisores.
Ecosistema microbiano y las enfermedades
No se ha demostrado plenamente la relación de los microbios con determinadas enfermedades. Sin embargo, es posible afirmar que la distensión intestinal, los cuadros diarreicos, las células cancerosas en el colon y el recto, así como algunas enfermedades metabólicas tienen que ver con los microbios.
Igualmente ocurre con el asma, las alergias y las afecciones del sistema nervioso central. También se han detectado afecciones como la depresión, el autismo, el Alzheimer, el Parkinson o la esclerosis múltiple.
No se sabe a ciencia cierta si las alteraciones en el ecosistema microbiano son la causa o la consecuencia de la enfermedad, pero se cree que ambos factores están relacionados. Aunque no se conoce a profundidad la relación con el cáncer, los microbios pueden colaborar con la efectividad de su tratamiento y mitigar o aumentar los efectos dañinos de tales tratamientos.
Cómo modificar el microbioma humano
La modificación de este sistema es difícil, por lo mucho que interactúan entre sí las células de los microbios y las células humanas. A pesar de ello, es posible lograr algunos cambios favorables para procurar su mejoría. Esto supone dos tipos de estrategias:
- Mediante reacomodo del sistema ya existente.
- Mediante nuevos microorganismos.
La primera de estas estrategias implica varios procedimientos:
- Cambios en la nutrición. Estos cambios son temporales, sólo serán permanentes si se diseña una dieta personalizada de parte de un especialista.
- Uso apropiado de los antibióticos. Para ello hay que evitar su administración sin prescripción facultativa. Sólo el médico conoce el antibiótico apropiado para cada colonia de microbios. Destruir microbios beneficiosos para la salud puede traer consecuencias adversas, como trastornos en enfermedades del sistema inmunológico, el padecimiento de los celíacos o la obesidad.
- Uso de prebióticos. Aunque no son digeribles por el organismo, ayudan al crecimiento de bacterias en el intestino que a su vez facilitan la digestión.
La inclusión de nuevos microorganismos
También en el ecosistema microbiano se ve la aparición de los siguientes nuevos microorganismos.
- Los probióticos. Son bacterias vivas que al ingerirse pueden eliminar bacterias contaminantes o producir moléculas de acción beneficiosa para el cuerpo humano.
- Simbióticos. La ingesta de un microorganismo probiótico junto a un prebiótico se traduce en un buen aporte nutricional.
- Trasplante de microbios. El ecosistema microbiano de un enfermo puede ser sustituido por el de un organismo sano mediante trasplantes, por cultivos o autotrasplantes.

El ecosistema microbiano y la prevención
El estudio y conocimiento de esos habitantes invisibles, que aquí se han tratado, se ha convertido en un reto para la medicina del presente y del futuro. Ello se debe a que se hace referencia a un sistema diferente para cada individuo y a los cambios que se producen a lo largo de su vida.
Esto implica hasta a los precisos estudios del genoma humano, pues estos deben tener en cuenta al sistema microbiano. Los resultados obtenidos se aplicarían en diagnósticos de precisión personalizados.
Hoy, gracias a la informática, se puede detectar estas interacciones que permitirán predecir futuras enfermedades de los pacientes. Igualmente se conocerán de antemano las posibles reacciones a determinados tratamientos y las estrategias más adecuadas de modificación microbiana.
No hay duda de que conocer con precisión la estructura y composición del ecosistema microbiano humano será un gran aporte para la medicina preventiva. De igual forma será muy provechosa en la diagnosis exitosa y en los tratamientos más adecuados para muchas enfermedades.
Referencias:
- Cell 171, December 14, 2017













0 comentarios